T7 SSB
T4 Polynucleotide Kinase
T4 Polynucleotide Kinase, Phosphatase (-)
T4 DNA Polymerase
MMLV reverse transcriptase
Vaccinia Topoisomerase I
Protein Expression Testing
AKTA FPLC (ion-exchange, IMAC)
Protein Purification Process Development
Non-Tagged Protein Purification
Complete Batch Records
Manufacturing Pilot Process
Scale-Up
Custom Protein Formulations
Tissue Culture (primary cells, adherent and suspension cell lines)
Viability Assays (fluorescent, colormetric)
Custom Competent Cell Preparation
(chemical, electrocompetent, 96 well
format)
(chemical, electrocompetent, 96 well
format)
Subcloning
Bacterial or Yeast Growth
Custom Protein Formulations
DpnI
You can also place an order by faxing a completed order form to 858-558-3740 or calling us at 858-558-3702.
Description:
DpnI restriction endonuclease is derived from the dpn1 gene from Diplococcus pneumoniae expressed in E. coli. This restriction enzyme is methylation-sensitive and requires the adenine within the restriction site is methylated at position N-6(1,2). DpnI prefers fully-adenomethylated dam sites; hemi-adenomethylated dam sites are cleaved by DpnI at 2% of the rate of fully methylated sites. Cleavage of eukaryotic genomic DNA may be blocked by overlapping CpG methylation.
Protein Uses:
Blunt-end PCR cloning
site-directed mutagenesis procedure
Methylation pattern recognition
FastCloning – sequence and ligation independent PCR cloning
Additional Information:
DpnI recognition sequence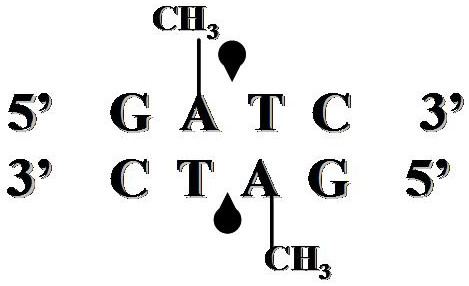

Available in high concentration (Cat no. 9002, 9006)
Formulated with BSA (Cat no. 9005, 9006) and without BSA (cat no. 9001, 9002)
Effect of various methyltransferases on DpnI recognition and activity:
Dam – necessary for recognition; DNA from Dam - strain will not be digested
Dcm – some recognition site overlap, no effect on activity
CpG – may overlap with recognition site and block activity
EcoKI – no recognition site overlap hence no effect on activity
EcoBI – some recognition site overlap; may affect activity
Dam – necessary for recognition; DNA from Dam - strain will not be digested
Dcm – some recognition site overlap, no effect on activity
CpG – may overlap with recognition site and block activity
EcoKI – no recognition site overlap hence no effect on activity
EcoBI – some recognition site overlap; may affect activity
